The Centrosome Size Project
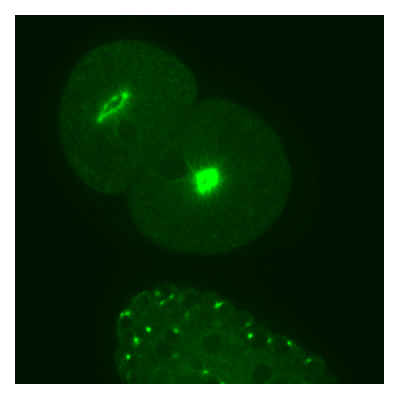
The centrosome is a dynamic organelle found in all animal cells that serves as a microtubule organizing center during cell division. Most of the centrosome components have been identified by genetic screens over the last decade, but little is known about how these components interact with each other to form a functional centrosome. In this project we set out to investigate the mechanism that regulates the size of the centrosome in the early C. elegans embryo. We used live-cell microscopy to monitor the growth dynamics of fluorescently labeled centrosomes. Various genetic perturbation techniques, particularly RNA interference and codon optimization, allowed us to infer the functions of individual proteins w.r.t. centrosome size. In order to analyse our time-lapse 3D movies, we have developed an automated image analysis tool, the centrosome tracker.
Our Matlab implementation of the automated image analysis tool can be downloaded here.
All centrosome movies, acquired between 2008 and 2011, are freely available for download here.
The entire collection is available via anonymous ftp from ftp://centsize.mpi-cbg.de.
References:
S. Jaensch 1, M. Decker 1, A. A. Hyman 1, E. W. Myers 2 (2010) “Automated tracking and analysis of centrosomes in early Caenorhabditis elegans embryos”, Bioinformatics 26, i13-i20. (PubMed)
M. Decker* 1, S. Jaensch* 1, A. Pozniakovsky 1, A. Zinke 1,
K. F. O'Connell 3, W. Zachariae 1, E. W. Myers 2, A. A. Hyman 1 (2011)
“Limiting amounts of centrosome material set centrosome size in C. elegans embryos“, Current Biology
(PubMed)
*These authors contributed equally.
1 Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany.
2 Howard Hughes Medical Institute Janelia Farm, Ashburn, Virginia, USA.
3 National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, Maryland, USA.